How many servers do we have?
We have three servers, one CPU server (Cyrus), one GPU server (Nautilus), and one Rstudio server (RSTUDIO).What is CYRUS?
 CYRUS is a CPU computing server intended to serve my graduate students, named after Cyrus the great,
who served humanity. This CentOS Linux server supports SSH and FTP services currently
running over 32 parallel processors and a total of 64GB RAM. I adminstrate CYRUS with help
of Benoit Forest, the computer technician of math department.
CYRUS is a CPU computing server intended to serve my graduate students, named after Cyrus the great,
who served humanity. This CentOS Linux server supports SSH and FTP services currently
running over 32 parallel processors and a total of 64GB RAM. I adminstrate CYRUS with help
of Benoit Forest, the computer technician of math department.
What is Nautilus?
 Nautilus is the Capitan Nemo's submarine which goes deep in oceans. It is also our GPU computing server, intended to serve my graduate students in training deep neural networks. This Ubuntu Linux server supports SSH and FTP services currently
running over 8 CPU that hosts a powerful Tesla K40 GPU server with 12 GB RAM and 2880 CUDA cores on one of its PCI 3.0 express slots. Khalid Laaziri adminstrates Nautilus.
To have access first you must ssh using "ssh -x geraduser@ssh.gerad.ca" and then "ssh -x nautilus.gerad.lan"
Nautilus is the Capitan Nemo's submarine which goes deep in oceans. It is also our GPU computing server, intended to serve my graduate students in training deep neural networks. This Ubuntu Linux server supports SSH and FTP services currently
running over 8 CPU that hosts a powerful Tesla K40 GPU server with 12 GB RAM and 2880 CUDA cores on one of its PCI 3.0 express slots. Khalid Laaziri adminstrates Nautilus.
To have access first you must ssh using "ssh -x geraduser@ssh.gerad.ca" and then "ssh -x nautilus.gerad.lan"You must "module load anaconda" and then "source activate keras-gpu" if you want to use keras with GPU acceleration, then run your python code using "python myfile.py". For more details contact Khalid. You may install what you like locally using "conda install".
What is RSTUDIO?
 RSTUDIO is a server that I use also as my office desktop. RSTUDIO on our server is equipped with a web interface that mimics the common RStudio Desktop, except that the computations are sent to this server. This server makes RSTUDIO available on cell phones!
RSTUDIO is a server that I use also as my office desktop. RSTUDIO on our server is equipped with a web interface that mimics the common RStudio Desktop, except that the computations are sent to this server. This server makes RSTUDIO available on cell phones!
CYRUS Hardware
Nautilus Hardware
RSTUDIO Hardware
How can I have access?
I must create you a username and send you a toy password. I strongly suggest to change your password as soon as you logged in the first time.How do I login?
You can connect to the server using ssh command. If you work on Microsoft Windows, you need to install Putty. Below I describe some basic hints supposing that the server is CYRUS. For Mac or Linux users the following command can be run directly from your terminalLOCAL$ ssh username@cyrus.mathappl.polymtl.ca
How to change password?
After you login on the serverCYRUS$ passwd username
How to copy files?
If you need to copy files from your computer to the server, or back from the server to your computer, you can use ftp. Install the ftp interface called fireftp on your mozilla firefox browser. The ftp server is cyrus.mathappl.polymtl.ca, see below for more details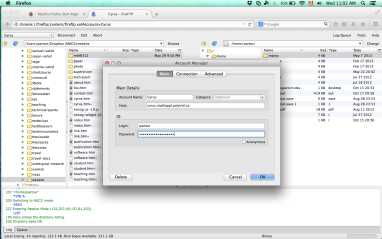
How to use R on CYRUS?
R is installed on the server. If you need some libraries, you have to install the libraries locally on your space on the server. First make a directory to install your local libraries in. CYRUS$ cd /home/username
CYRUS$ mkdir Rlibs
CYRUS$ R
R> install.packages( "mylibrary", lib = "/home/username/Rlibs" )
R> library( "mylibrary", lib.loc = "/home/username/Rlibs/" )